CORONAVIRUS Library
Outbreak of coronavirus disease (COVID-19) was first reported from Wuhan, China, on 31 December 2019.
Coronaviruses (CoV) are a large family of viruses that cause illness ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS-CoV). A novel coronavirus (nCoV) is a new strain that has not been previously identified in humans.
Coronaviruses are zoonotic, meaning they are transmitted between animals and people. Detailed investigations found that SARS-CoV was transmitted from civet cats to humans and MERS-CoV from dromedary camels to humans. Several known coronaviruses are circulating in animals that have not yet infected humans.
Chromis Virtual Screening Methodology
Swiss Prot Protein Targets and PDB X Ray Structure Search: ACE2, PLpro , 3CLP
Training Sets ChEMBL 25, PubMed, Current Patent Literature (CAS, Integrity)
Machine Learning Data Curation:
a) KNIME/ RDKit , kNN classifier, Distance in BitVector Cosine Space, FCFP12 (10,240 bit) fingerprints
b) Hybrid 2D QSAR/Fingerprint Model Kernel Chemical Classification/Regression (kcc)
3D Shape Similarity
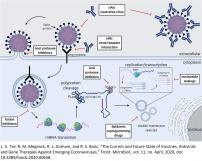
Virtual Screening :
APF®MolSoft , Lam et al J. Comp Aided. Mol. Design (2017, 2018 & 2019); APF Totrov Chem Biol Drug Des. (2008)
Structure-Based (Ligands, Fragments, Covalent Fragments) Docking / Virtual Screening
a) Multiple Receptor Conformation (MRC) 4D Docking; ICM Pro MolSoft , Bottegoni et al (2009) J. Med. Chem. 52:397
b) Ligand Biased Ensemble receptor Docking ( LigBEnD ); ICM Pro MolSoft , Lam et al J. Comp Aided. Mol. Design (
REOS, MedChem & PAINS Filters – Removal of reactive, toxic, promiscuous, and other undesirable structural motifs
Diversity Picking (Tanimoto): RDKit implementation of the MaxMin algorithm Ashton, M. et. al., Quant. Struct. Act. Relat ., 2002, 21, 598 604.
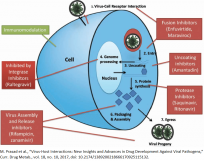
2. Nucleic Acids Research 2019, 47(12):6538 6550, Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Jia Z, Yan L, Ren Z, […], Rao Z. doi.org:10.1093/nar/gkz409
3. Nat. Commun . 2019, 10:2342, Structure of the SARS CoV nsp12 polymerase bound to nsp7 and nsp8 co factors. Kirchdoerfer RN, Ward AB. doi.org:10.1038/s41467 019 10280 3
4. J. Biol. Chem. 2004, 279: 17996 18007, ACE2 X ray structures reveal a large hinge bending motion important for inhibitor binding and catalysis. Towler P, Staker B, Prasad SG, […], Pantoliano MW. doi:10.1074/jbc.M311191200
5. Expert Rev. Anti. Infect. Ther . 2006, 4(2):291 302, Potential antivirals and antiviral strategies against SARS coronavirus infections. De Clercq E. doi:10.1586/14787210.4.2.291
6. FEBS J. 2014, 281(18):4085 4096, From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. Hilgenfeld R. doi:10.1111/febs.12936
7. Bioorg . Med. Chem. 2015, 23(4):876 890. Fused ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Shimamoto Y, Hattori Y, Kobayashi K, […], Akaji K. doi : 10.1016/j.bmc.2014.12.028
8. Biopolymers 2016,106(4):391 403. Structural basis for the development of SARS 3CL protease inhibitors from a peptide mimic to an aza decaline scaffold. Teruya K, Hattori Y, Shimamoto Y, […], Akaji K. doi : 10.1002/bip.22773
9. Bioorg . Med. Chem. 2019, 27(2):425 435. Evaluation of a non prime site substituent and warheads combined with a decahydroisoquinolin scaffold as a SARS 3CL protease inhibitor. Ohnishi K, Hattori Y, Kobayashi K, Akaji K. doi : 10.1016/j.bmc.2018.12.019